The rise of the machines
Over the past several years, I’ve stumbled into two fields of research which, although differing in their subject matter, are united by the recent entrance of computational methods into domains that were previously dominated by manual modes of analysis.
One of these two fields is computational social science. This is the broad category into which my all-but-examined PhD thesis, and most of the posts on this blog, could be placed. Of course, my work, which focuses on the application of text analytics to communication studies, occupies a tiny niche within this larger field, which encompasses the use of all kinds of computational methods to the study of social phenomena, including network science and social simulations using agent-based models.
The other field to which I can claim some degree of membership is the digital humanities. This tag is one way to describe what I do on my other blog, in which I have drawn on digitised maps and newspapers to produce visualisations that explore the history of a suburban environment. History is just one field of the humanities that has been opened up to new possibilities through the extensive digitisation of source materials. Scholars of literature have joined the party as well, while those operating in the GLAM sector, consisting of galleries, libraries, and museums, can now use digital platforms to manage and share their collections.
Though often lumped together in the same faculty (or indeed the same school at my last university), the humanities and social sciences don’t generally have a lot to do with one another. They do, however, have commonalities that lend themselves to similar computational methods. One is the study of text, often in large quantities. Within the social sciences, communication scholars analyse (st)reams of news texts and social media feeds; qualitative sociologists often amass hours of interview transcripts; political scientists analyse speeches and parliamentary transcripts. In the humanities, scholars of history and literature now have at their disposal huge archives of digitised novels, journals, newspapers, and government records. Of course, there is nothing compelling scholars in these fields to analyse all of the available data at once. They are free to sample or cherry-pick as they please. But any scholar wishing to work with large textual datasets in their totality can hardly afford to ignore computational tools like topic models, named entity extraction, sentiment analysis, and domain-specific dictionaries such as LIWC.
Another analytical method that is applicable within both the social sciences and humanities is the study of networks. Sociologists and historians have long used networks to represent and analyse social ties. More recently, social media platforms have hard-wired the logic of networks into social relationships, explicitly representing all of us as nodes tied to one another by virtue of our acquaintances, interactions, and browsing histories. Accordingly, it is becoming increasingly difficult to study social phenomena without invoking the concept of networks, if not also the science of network analysis. By no means restricted to real-world phenomena, network analysis techniques are equally applicable to fictitious societies, whether those in Game of Thrones or the plays of Shakespeare. As with the analysis of text, network analysis doesn’t have to be done with a computer; but as the size of a network grows, so too does the need for computational tools.
Besides an interest in certain computational methods, something else that the digital humanities and computational social sciences have in common is that many of the scholars trying to break into these fields are doing so without any training in data science or computer programming. This is especially the case in the humanities, where traditionally, quantitative methods have been virtually taboo. In some branches of the social sciences, quantitative methods are well established, but these are more likely to take the form of spreadsheets and statistical tests rather than machine learning and network analyses.
(Of course, the story is very different for those researchers entering these fields from the other direction, such as the many physicists, mathematicians and computer scientists who have somehow claimed the territory of computational social science. What these researchers lack, although many of them seem not to realise it, is any training in understanding the actual substance of what they are studying.)
The bottom line is that for many researchers, embracing the new opportunities presented by the digital humanities and computational social sciences means learning skills that are totally outside of their disciplinary training. At a minimum, researchers hoping to use computational methods in these fields will need to learn how to calculate statistics and use spreadsheets, and perhaps also how to use network visualisation software like Gephi or text-cleaning software like OpenRefine. Increasingly though, getting your hands dirty in computational social science or the computational end of the digital humanities 1 means learning something far worse — something that is commonly denoted by a certain four-letter word.
The C-word
Applying computational methods in the social sciences and humanities means more than just performing a statistical test or tabulating data. Typically, it involves some combination of obtaining data from online sources via APIs or scraping tools, cleaning and refining such data to get it ready for analysis, and performing a long sequence of analytical operations which draw on many different inputs and produce just as many new outputs, perhaps across a variety of numeric, textual and graphical formats. In addition, all of these steps need to be retraceable and repeatable, not only so you can build and update the sequence in an iterative fashion, but also so that others can assess and replicate your findings.
In other words, many of the methods used within computational social science and digital humanities are not merely computational; they are programmatic. They can only be done by executing a long and possibly complex sequence of operations. For a computer to execute them, these operations need to be embedded in a series of unambiguous, carefully assembled instructions, otherwise known as code.
To my observation, which I admit is far from extensive, coding skills are a something of a dividing line within the humanities and social sciences. In a practical sense, coding is the gateway to all kinds of tools and methods that otherwise seem out of reach. Even some fairly basic skills in R or Python can transform the way you manipulate data while also providing access to specialist tools like topic modelling or natural language processing. Consequently, coding divides and defines practitioners in these fields. Regardless of your other research skills or interests, if you can code, you are likely to pursue different directions, whether by choice or because you’re “that person” who non-coding colleagues may turn to or lean on when they want an edge in their next project.
But learning to code is a big deal — or at least, it’s made out to be. While I have been writing this post, “learn to code” has even surfaced as a meme used to mock and harass journalists who have been laid off, as if their careers and society at large would be better served by computer programming than by news reporting. I expect that many of the people deploying this meme would happily also use it against scholars in the humanities and social sciences.
Undoubtedly, learning to code demands a considerable investment in time and energy that many researchers simply cannot justify, especially if their existing skillset is serving them well. On the other hand, this investment might seem like a no-brainer for a more adventurous researcher who has time on their side. However, even for someone in this position, the prospect of learning to code may seem daunting, particularly in the absence of any direct access to training or support. From a distance, learning to code might seem too alien, or just too hard, to devote time that might otherwise be spent getting on with things that you already know how to do.
I’m talking from my own experience here. In the early stages of my PhD, I faced this very dilemma. After a circuitous route to commencement (which included enrolling initially with a totally different project in a totally different school), I began under the assumption that I would explore the nexus between text analytics, data visualisation and communication studies by taking advantage of graphically interfaced tools like Excel, Tableau, Leximancer, and Gephi. While possessing some technical aptitude, I had no formal training at all in computer science, and the closest I had come to coding was fiddling with HTML and CCS on my blogs. I figured I’d focus more on the social science content than on the details of the methods, and declared that I would teach myself to code only if and when I absolutely had to.
Four years later, I found myself with a data analysis skillset that I never expected to have. Among other things, I’ve become reasonably proficient at doing things like acquiring and processing textual data, generating and comparing topic models, fusing the outputs with contextual variables like geography and authorship, and using the outputs to create networks of documents or discourse actors. And I can do it all in a programmatic way, creating workflows that can be templated and adapted to new datasets.
But here’s the thing: I’ve done virtually all of it without a line of code. After a brief flirtation with R in the second year of my PhD, I barely touched it again until recently, and only in order to create better data visualisations. I’ve attended one or two workshops about Jupyter notebooks, but otherwise I have not touched Python.
This is all possible because I found a third way — a path between the seemingly opposing options of coding and graphical user interface (GUI) tools. That third way is called Knime (or KNIME if you want to honour the official capitalisation), and while I’ve mentioned it often enough in my blog posts, I’ve never discussed it at any length. With this post, I’d finally like to give Knime the credit it s due, and explain why I think it deserves a more prominent place within the ecosystems of computational social science and digital humanities.
Coding without code
Whenever I explain that I use a tool called “Knime — K-N-I-M-E”, I am usually met with a blank stare. Sometimes, that stare is sometimes followed politely with “I must look that up”. In my experience, almost no-one has heard of Knime. And after I’ve mentioned it, if someone ever does have cause to refer to “that tool” that I use, they instinctively search for the most creative way to pronounce it — usually ‘Kuh-nime’, ‘Kay-neem’, or some similar variation. Perhaps they are following the unfortunate example set by Gnome, the Linux (Leenux? Lyenix?) distribution that for technical reasons that make sense only within the Linux universe is supposed to be pronounced with a hard G, even though many users stubbornly resist. 2 Or perhaps the official pronunciation of Knime, which has a silent K and rhymes with, erm, rhyme, just sounds too strange or non-technical. (I don’t know what a kayneem could be, but it sounds very sciency.) Admittedly, this is a world of confusion that users of R don’t have to deal with.
In any case, the silent K, in combination with the non-silent N, denotes the German university city of Konstanz, where the Konstanz Information Miner was first developed (I can only assume that the more logical and personable acronym, KIM, was already taken). The developers, who are now based in Zurich, Switzerland, still write the name in all-capital letters, but as you can already see, I prefer to write it in title case, mostly because it looks less shouty, but also because many acronyms ultimately end up being written that way anyhow. 3 And I’d be willing to bet that far more people would pronounce Knime correctly if it were capitalised in this way.
Knime is an open-source data analytics platform written in Java (the programming language, not the island). Originally developed for use in the pharmaceuticals industry, it has since evolved into a more general-purpose platform, much like R or Python. Within the university sector, its use is still fairly limited, and, I suspect, concentrated in areas such as pharmacology and biological science. Judging from its Wikipedia page, Knime is used primarily within industry, including banks, telcos and consulting firms.
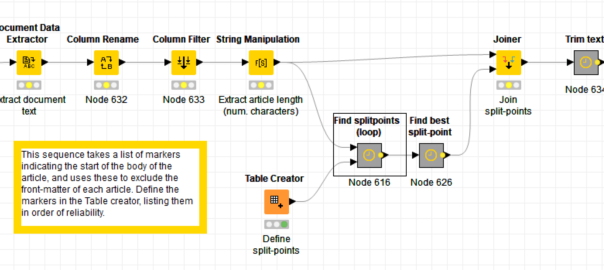
The thing that makes Knime different from most other data analytics tools — and this is quite possibly why it deters academic researches while attracting industry professionals — is that it uses a graphical user interface. Instead of writing lines of code, the user builds data pipelines, or workflows, out of pre-defined nodes. Here are some excerpts from my own workflow for removing the unwanted text at the start of journal articles:
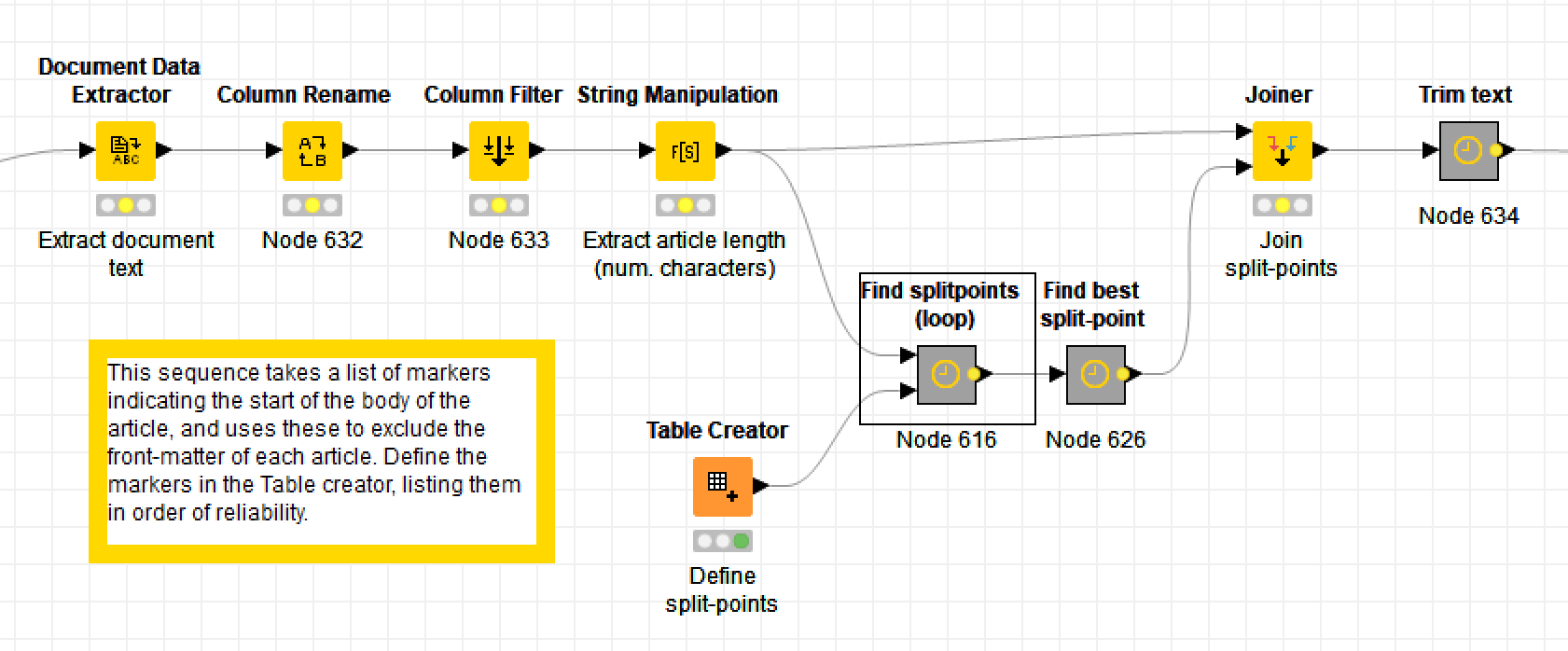
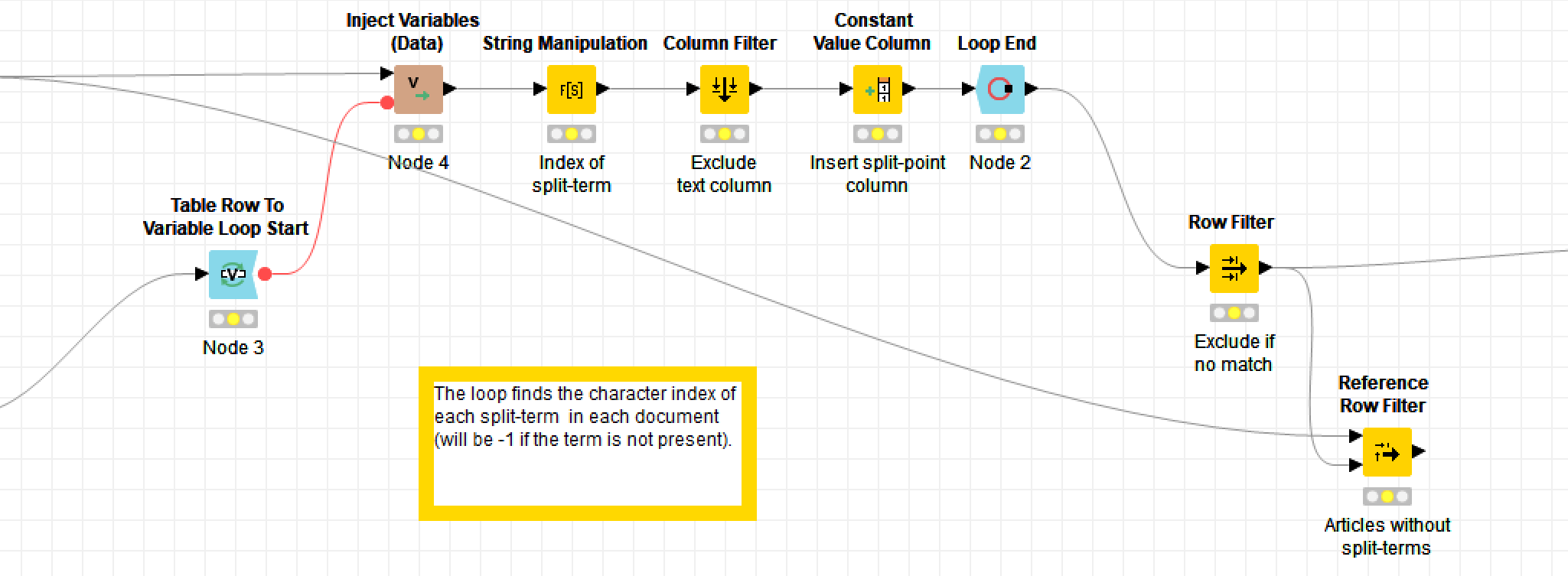
In these excerpts, the bold labels above the coloured nodes are the names of the nodes: they describe what generic operations each node performs. The grey boxes are metanodes, which, as the name suggests, are simply nodes made up of more nodes. The bold labels above these are user-defined, as are the optional descriptions beneath the ordinary nodes (by default, the nodes are just given an ID number).
Obviously, there is much that you can’t tell about the workflow just by from looking at the screenshots, such as which columns are being renamed or filtered, or what string manipulation formulas I’ve used (string manipulation is one task for which you’ll still need to write code-like snippets). These details are specified within the node configurations, which you access by double-clicking a node. But while the details are hidden, the broader flow of events is plain to see. If you understand the basic logic of a flowchart, you know enough to build a Knime workflow — provided, of course, that you also know what operations you want to perform on your data.
The variety of such operations on offer in Knime’s node repository is impressive, ranging from basic data manipulations right through to data clustering, text processing, statistical modelling and machine learning. In short, it covers many of the things that you might otherwise rely on R, Python, Matlab or SPSS to do. And for the things that Knime can’t do, there are nodes enabling seamless integration of R, Python and other platforms.
And as far as I know, there is no limit to how big and complex a workflow can be. As a workflow grows in complexity, you can simply group sets of nodes into metanodes, which can be nested many layers deep.
So, what may look like a GUI toy for people who are not serious enough to code (and I have no doubt that Knime is dismissed this way by some people), is in fact a fully-fledged platform for analysing data in a programmatic fashion. It’s like coding, but without the code. Or you could say that Knime substitutes one form of coding — if we want to reclaim the silent K, perhaps we could call it ‘koding’ — for another. Instead of using a code based on a vocabulary of commands that must be parsed via a strict and terse syntax, Knime uses a library of functions that are configured through interactive menus and connected through an intuitive visual grammar based on pipelines and data flows. It might look more like plumbing than programming, but it achieves much the same thing.
Coding vs Koding
This visual coding approach comes with both advantages and disadvantages, some obvious, some more subtle. The most obvious advantage of Knime is that it is more intuitive than conventional coding. If you understand what each node does, then you can build a workflow. All you need to do is configure the nodes (which is done through plain English, point-and-click menus) and join the nodes together in the right order. There is no special syntax to learn, no brackets to parse, no list of command names and parameters to internalise. As a result, the path from being a beginner to producing results is much faster than with traditional coding, because you can focus on the content of what you are doing rather than also having to learn a whole new grammar and vocabulary. All that time and energy that would otherwise be spent searching Google and Stack Overflow for things as trivial as “how to rename a column in R” is freed up, and can be allocated to the substance of what you are actually trying to achieve.

The visual grammar of Knime also makes the logic and structure of a workflow more transparent. Whereas code presents as strictly linear (it’s just one line after another), a Knime workflow is more akin to a flowchart or concept diagram, showing explicitly how multiple inputs and parallel operations intertwine with one another. This means that when you are working with structurally complex procedures, you don’t need to draw a map to trace the connections, because the map is already in front of you.
Even better, the map is interactive. You can inspect the outputs of any node just by clicking on it. Whereas tools like R Studio also allow you to inspect interim outputs without much hassle, they typically do so by providing a list of named objects that is visually separate from the script itself. In Knime, you are able to interrogate the workflow more directly, which I find to be especially useful when you are still getting a feel for what the nodes do to the incoming data.
So, Knime is easier than code to learn, and makes complex sequences easier to trace and inspect. But these benefits come at a price. One downside of Knime workflows is that they cannot be shared and moved around as easily as code. The abstractness of code means that it is information-dense. Each operation is encoded in few bytes of information — a handful of ASCII characters that can be copied and pasted with perfect fidelity from a text editor to a web browser to an online repository. A Knime workflow looks simple because most of the complexity is hidden. Without opening the workflow in Knime itself, you have no way of knowing how the nodes are configured. In fact, without Knime itself, you can’t inspect a workflow at all. Unlike with code, you can’t paste a Knime workflow into a text editor or a web form. Even porting content from one Knime workspace to another is harder than it should be. And on top of all that, the graphical interface comes with overheads: more memory and processing power that could otherwise go towards executing your code, and more ways for things to go wrong, potentially causing your software or operating system to crash (that said, I have found Knime to be remarkably stable and robust, and I’ve had R Studio crash on me plenty of times).
Aside from the way in which you get things done, there is also the question of what things you can actually do. As I mentioned earlier, Knime has an impressive library of nodes from which to construct workflows. But the selection is not as great as it would be if you are using R or Python. For example, Knime has a node that performs topic modelling, but it offers only a minimally configurable implementation of LDA. If you want to use other topic modelling algorithms, such as the structural topic model or correlated topic model, you’ll have to use another platform, such as R. (As a consolation, however, you can use Knime’s R integration features to embed an R script within your Knime workflow.)
The bottom line is that Knime is just another way of doing may of the things that you might otherwise do with R, Python, or other data analysis platforms. It’s easier to learn and to use than code-based platforms, but is in certain respects less flexible and less convenient. It has some pros, it has some cons. From a potential user’s point of view, the question is which of these pros and cons really matter?
EDIT, 22 March 2019: Something I did not mention in the above discussion, but which for some people would be an absolute dealbreaker, is that if you want to turn a Knime workflow into a web interface, the product stops being free and starts being expensive. You’ll need KNIME Server, which is a commercial extension to the open-source component of Knime. Having recently gotten one of my own workflows to a point where it is begging to be turned into a web interface, this is a sobering realisation.
Data analytics for the rest of us
As I’ve alluded to already, I doubt that any self-respecting computer scientist would be caught dead using Knime. He or she did not spend years learning to do things the hard way — the proper way — just to turn around and use a tool with a point-and-click interface. And they might have a point. If you are already proficient in R or Python, there is probably not much reason to use Knime. 4
But, as I discussed at the start of this post, most scholars in the humanities and social sciences are not already proficient in R or Python. For many people in these fields, the mere idea of learning to code in such platforms is terrifying. Or even if it isn’t, it can seem like too big and too risky an investment of time to be worthwhile.
These are the people, I think, who have the most to gain from using Knime. The reason is that Knime presents less of a barrier to entry than coding, and yet enables most of the same outcomes to be achieved. Since the average humanities or social science scholar is unlikely to be concerned about the elegance and purity of code, or the kudos that comes with being able to wield it, why would he or she invest time learning to code if the same outcome could be achieved by simpler means?
My impression (again, gained from limited experience) is that many people in the humanities and social sciences feel that learning to code is the only available avenue if they want to do any serious data analysis. As a result, many opt out altogether, or pin their hopes instead on customised tools or services (sometimes under the banner of ‘infrastructure’) that enable them to do at least something that buys them a spot on the DH or CSS bandwagon. Perhaps if middle-road alternatives like Knime were visible instead, more people in these fields might take the plunge and develop some new data analysis skills.
This isn’t to say that everyone in these fields can or should learn such skills. Knime makes data analysis easier, but not easy. And there will always be plenty of important scholarship to be done that doesn’t require big data or computational methods. Nor am I suggesting that Knime is the only sensible tool for scholars in these fields to use. If your likely collaborators all use another platform, then you might as well learn to use it as well. Or if having access to the most obscure and cutting-edge packages is a priority, then you might be frustrated if you limit yourself to Knime.

Importantly, however, using Knime doesn’t mean you can’t learn to use R or Python as well. Indeed, I’d be willing to bet that learning to use those platforms will be a whole lot easier if you have spent some time with Knime first. Through Knime you can learn the basics of data manipulation and analysis: switching to another platform is then just a matter of translating that knowledge into another language. In my case, I thought initially that Knime would serve as a gateway drug to R or Python. But four years on, I still have little incentive to use other platforms, except to fill in the gaps in Knime’s capability, such as data visualisation. 5
This all begs an obvious question: if Knime is so well suited to folk in the digital social sciences and humanities, why has it not been more widely adopted in these fields? My best guess is that it is because scholars in these fields have followed the lead of researchers from computer science departments, who would have little interest in, or even any knowledge of, a tool like Knime. Naturally, any computer scientist who reaches out to, or responds to a call from, their colleagues in the HASS faculty will share the knowledge and methods that they themselves have learned. But what is best for computer scientists is not necessarily what is best for the rest of us.
I could be proven wrong. Perhaps code-based platforms will continue to rule within the academy, whether for good reasons or bad. Perhaps in the not-too-distant future, even students embarking on humanities degrees will already be fluent coders by the time they get to university. Perhaps, but perhaps not. Whatever the case, I feel like my own experience with Knime is worth sharing, and if it does deserve a place in the digital humanities and/or computational social sciences, I want to do what I can to spread the good word.
Show us your kode!
A common question asked in response to demonstrations of computational analyses in the social sciences and humanities — just as in computer science — is “where can I see your code?” The practices of sharing code in academic research is an admirable one. Not only does it allow other researchers to scrutinise your analysis; it also enables others to learn from and build upon your work. And as I mentioned earlier, one of the advantages of code is that it can be shared so easily.
As yet, I’ve not offered any of my own Knime workflows for others to peruse or scrutinise. The main reason is that my workflows to this point have not been in a fit state to share, being the result of me learning by the seat of my pants most of the time (something that is easy to do with an intuitive tool like Knime!). Among other things, I’ve learned the hard way that you should always annotate your code. And as with conventional code, or perhaps even more so, there are many different ways to set out the components of a Knime workflow, some of which make for much easier comprehension than others.
Having nurtured my Knime workflows to a point where they are likely to be decipherable by others, I now plan to start sharing them. As a first offering, I have placed on Github the workflow that I used to produce the analysis in my last post analysing what I downloaded and cited during my PhD candidature. Using annotations, I’ve done my best to document each step, explaining what is going on and what options the user may want or need to review. One thing the workflow doesn’t include is the PDFs that were the subject of my analysis. Aside from the collection being quite large (about 160MB), I suspect that I couldn’t legally distribute them all anyhow. Instead, I’ve included some pre-parsed sample data, and the processed text of my full collection so that you can explore the topic modelling and analysis nodes.
The ideal outcome of me doing this would be for someone to use this workflow to analyse their own collection of downloaded literature. But even if you don’t have such a collection to analyse, you may wish to explore the workflow just to see how Knime can be used for this kind of analysis. Included in the workflow are sequences of nodes that do the following things:
- Extracting metadata from file names (this step uses logs of regex, which you’ll still need to learn if you want to do any serious text processing in Knime)
- Removing ‘boilerplate’ text such as publisher information, from the front of journal articles, and references from the end.
- Text preprocessing, including filtering of named entities, stop-words, URLs and numbers. This includes my own experimental method for standardising plurals and other word-tense variations (I don’t like stemming as it tends to be too aggressive), as well as my own method for automatically tagging notable 2-grams and 3-grams.
- Generating LDA topic models, including my own methods for comparing pairs of candidate models (I will add a method for multi-model comparisons in the near future), and for clustering similar topics according to their terms or document allocations.
- Creating a similarity network of the articles based on their topics. You’ll need to load the outputs into Gephi to visualise the networks. (Knime has its own network analysis nodes, but I have not had time to familiarise myself with them.)
- Extracting citations from a larger document (e.g. your thesis) and reconciling them with the downloaded documents (assuming they have associated author and year metadata).
I’ve done my best to wrap up these and other discrete functions into metanodes that can be transplanted and adapted to other workflows. In future, I might offer them as separate workflows altogether, alongside more purpose-specific mega-workflows like this one (already, I’m realising that the best way to do this would be to make more use of Knime’s ‘workflow groups’ feature). With any luck, I will also find time to update the workflows that I share, as I tend to improve upon them with each new application (and in some cases, there is still a lot of room for improvement!).
I’ll be really interested to see how things pan out in the digital humanities and computational social sciences over the coming years. Will the divide between coders and non-coders persist, and perhaps become even more entrenched? Will coding become such a common skill that the average historian will think nothing of switching back and forth between Python, Latin and ancient Greek? Or will tools like Knime give rise to a new cohort of koders who opt for a shorter, more pragmatic pathway to computational methods? I’ll be interested too to see if I am still using Knime in a few years, or if I will have migrated to a fully code-based environment.
And finally, I’d love to hear other people’s experiences and thoughts about using Knime in the social sciences and humanities. If you have something to share in this regard, please don’t hesitate to leave a comment or get in touch.
Notes:
- There really is a difference between the digital in digital humanities and the computational in computational social science. Much of what falls under the digital humanities pertains only to the use and curation of digitised resources, rather than the use of computational methods to analyse these resources. Computational social science, on the other hand, seems to be driven primarily by the desire to analyse social data through computational methods. ↩
- One commenter on this Reddit thread said that (s)he “pronounce[s] gnome like the garden gnome and GNU like the animal of the same name precisely because stallman doesn’t want me to. ↩
- The word laser, whose letters stand for ‘light amplification by stimulated emission of radiation’, is a classic example. But there is precedent for adopting the convention more generally. Some style guides, such as that of the Guardian, specify that if an acronym is pronounced like a word (instead of a series of letters), it should be written like one. ↩
- It is possible that certain things are easier to do in Knime even if you are proficient in R or Python, but I don’t have the necessary experience in those platforms to know. Perhaps I’ll find out if I ever try to reproduce one of my Knime workflows in R. ↩
- Knime does have some useful data visualisation tools, but nothing of the calibre of ggplot and other packages available in R. ↩
This approach can be used for analysing the news about the stock markets. It can be harnessed for speculative trading. Stock market is mostly driven by speculation and being on top of them would give the investor a quite an edge.